Functional Genetics and Genomics
Team Members
Derek Morris
Richard Anney
Ziarah Hawi
Elaine Kenny
Matthew Hill
The molecular genetics research within the Neuropsychiatric Genetics Research Group can be divided into three strands:
Sequencing – Detection of sequence and structural variation in disease and control samples using low throughput capillary sequencing or ultra-high throughput next generation sequencing.
Genotyping – Assaying genetic variation a various types of genetic markers (e.g. SNPs) using low throughput platforms (e.g. Taqman) or high throughput genome-wide arrays.
Analysis of Functional Variants – Characterization of the impact of specific mutations/polymorphisms on gene function using a range of molecular, cellular and bioinformatics tools.
Translational Research
TrinSeq, the Trinity Genome Sequencing Laboratory is located in the Institute of Molecular Medicine (IMM), Trinity College Dublin. The laboratory now houses an Illumina MiSeq, a benchtop next generation sequencing platform that can undertake a range of ultra-high throughput genomics applications. This is a core facility supported by full-time research staff available to all academic and industry-based researchers in Ireland.
MiSeq Technology 
This ultra high-throughput sequencing system enables a wide range of genetic analysis applications including de novo sequencing of novel genomes, resequencing of whole genomes or target regions (e.g. candidate disease genes), transcriptome profiling and the study of DNA regulatory mechanisms. The MiSeq system is powered by Illumina sequencing technology, which uses a massively parallel sequencing-by-synthesis approach to generate billions of bases of high-quality DNA sequence per run.
Applications
|
DNA Sequencing |
Transcriptome analysis |
Gene Regulation and Control |
|
|
Genome-wide analysis of
|
Core Facility
The Trinity Genome Sequencing Laboratory provides the following assistance to researchers interested in undertaking a project using the Illumina MiSeq Benchtop Sequencer:
- Advice on study design and costs
- Advice and guidance on sample library preparation
- Access to automated liquid handling for large scale projects
- Sequencing and primary data analysis
- Advice and guidance on secondary data analysis
How To Get Started
For online information on how to set up your MiSeq sample prep and sequence run: http://support.illumina.com/sequencing/sequencing_instruments/miseq/training.ilmn
For more information on MiSeq reagent kits:
http://support.illumina.com/sequencing/sequencing_kits/miseq_reagent_kit.ilmn
For a list of publications using the Illumina Sequencing Technology go to: Publication Reviews | Highlights of recently published research (illumina.com)
For project enquiries/project quotes please contact: Dr Elaine Kenny
The Trinity Genome Sequencing Laboratory was established with funding from Science Foundation Ireland through a grant awarded to Dr. Derek Morris.
High throughput genotyping
In collaboration with Dr. Sean Ennis at University College Dublin as part of the Autism Genome Project, the group currently operates the Illumina BeadArray System, a whole genome genotyping platform that supports integrated single nucleotide polymorphism (SNP) and copy number variation (CNV) analysis. Using the Human1M-duo Infinium HD Beadchip, samples can be assayed genome-wide for over one million DNA markers.
Low throughput genotyping
There are a range of low throughput genotyping techniques available in our laboratory. These techniques can be applied to different types of genetic variants, e.g. SNPs, insertion/deletion polymorphisms (indels), microsatellite markers and variable number tandem repeats (VNTRs).
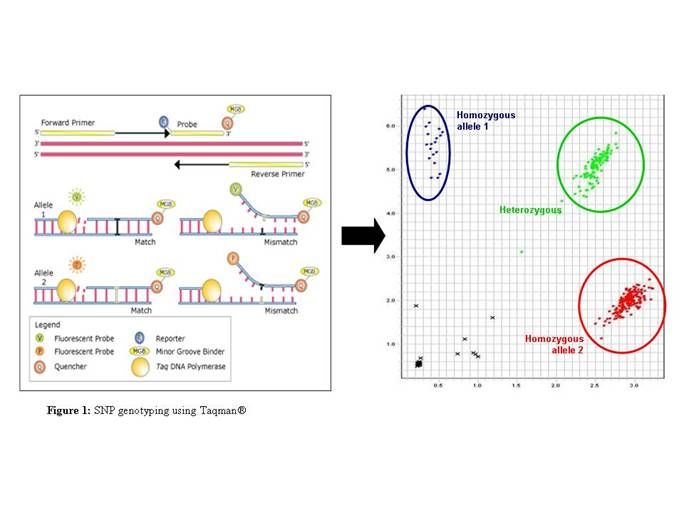
SNPs can be assayed using either the Taqman® genotyping method (see figure 1) on a 7900HT Real-Time PCR System (Applied Biosystems) or the SNaPshot® genotyping method on a 3130xl Genetic Analyzer (Applied Biosystems). We can increase the throughput of both methods by using robotics and operating in a 384-well format. Indels, microsatellite markers and VNTRs can be genotyped by size separation of fluorescently labelled PCR product on the 3130xl DNA Analyzer.
Team Members
Richard Anney
Matthew Hill
Summary Of Research
Many research programmes identify variation in the genome that is associated or linked to a trait. It is important to translate these findings into tool-sets that can assist in early diagnosis, intervention and treatment strategies. We are using a range of molecular, cellular and bioinformatics tools to examine the intermediate step in this process, namely the characterisation of the "functional" impact of statistically associated variants.
The functional elucidation of a statistically association variant will further our understanding of disease aetiology and provide evidence to support diagnostic and therapeutic strategies. In the Functional Variation Research Group we use a range of wet-lab and bioinformatics approaches to investigate common functional diversity in human genes. In the laboratory we apply a range of cell-based techniques include Allele Expression Imbalance (AEI) and gene-reporter technology. AEI of the gene message and of Immunoprecipitated DNA is a powerful quantitative technique for investigating cis-acting variation while minimizing confounding trans-acting factors. Our work overlaps with that of the Bioinformatics Research Team, where we examine is silico approaches to characterise the putative functionality of association at the SNP, gene and pathway level.

Collaborations
International Multicentre ADHD Genetics Programme
Knight Laboratory, Wellcome Trust Centre For Human Genetics, Oxford
ADHD Molecular Genetics Network - Functional Genetics Working Group
Autism Genome Project
Funding
Health Research Board Project Grant (2007-2008): Functional Significance of Common Genetic Diversity: the role of common haplotypes on the expression of genes associated with ADHD
Principle Investigator: Richard Anney
Neurodevelopmental disorders, including autism and schizophrenia, impair brain function and lead to behavioral, social and cognitive impairments. Recent advances in our ability to study the human genome have prompted significant research breakthroughs, including by researchers at the Neuropsychiatric Genetics Lab at TCD, in identifying susceptibility genes for autism and schizophrenia. Although there have been substantial progress in understanding the genetics of these disorders, their neurobiology remains obscure.
In the translational research lab, we study the biological function of susceptibility genes for neurodevelopmental disorders identified with genetic screens. These genes can be studied in vitro and in vivo using transgenic animals. These models play a critical role in helping to understand the neurobiology that leads to the expression of neurodevelopmental disorders, for improving diagnosis and leading to new treatments.
Collaborations
Dept. of Brain and Cognitive Science, Massachusetts Institute of Technology, Cambridge MA USA.
Funding
Marie Curie IRG 248284. Researcher Dr. Daniela Tropea
Lead Researcher: Dr Daniela Tropea, Lecturer in functional genomics
Research Assistant: Dr Ines Molinos
