Trinity Scientists Identify A New Type of MRSA
Posted on: 03 June 2011
A distinctly new type of methicillin-resistant Staphylococcus aureus (MRSA) has been discovered in patients in Irish hospitals by Trinity scientists according to research just published in the journal Antimicrobial Agents and Chemotherapy. The findings provide significant insights into how new MRSA strains emerge and highlight the potential for the transmission of infectious agents from animals to humans. The Dublin Dental University Hospital Microbiology Unit funded the study.
MRSA is a significant cause of hospital and community-acquired infection worldwide. MRSA strains are characterised by the presence of a mobile DNA cassette (known as SCCmec) encoding genes that confer resistance to beta-lactam antibiotics including methicillin and recombinase genes that allow the cassette to transfer into methicillin-susceptible S. aureus (MSSA).
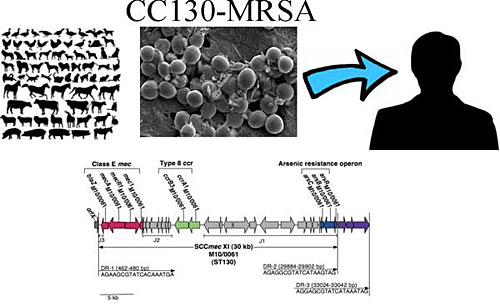
Professor David Coleman, Dr Anna Shore and Ms Emily Deasy at Trinity’s School of Dental Science, Division of Oral Biosciences, together with the National MRSA Reference Laboratory at St. James’s Hospital, Dublin and collaborators at the University of Dresden and Alere Technologies in Germany identified the new MRSA strain using high throughput DNA microarray screening. Complete genome sequencing of one of the new MRSA isolates revealed that this strain is distinctly different to previously described MRSA from anywhere in the world. This strain carries a new type of SCCmec that is very different to any described previously in MRSA or in any other organism. Consequently, the new strain is not detected as MRSA by routine conventional and real-time DNA-based polymerase chain reaction (PCR) assays commonly used to screen patients for MRSA. The MRSA strain was found to belong to the genetic lineage clonal complex 130 (CC130), which has previously only been associated with MSSA from cows and other animals, but not humans, strongly suggesting that the new MRSA originated in animals.
During the publication process, the TCD researchers became aware that a consortium of researchers led by the University of Cambridge and the Wellcome Trust Sanger Institute in the United Kingdom in an independent study had identified bovine MRSA harbouring an identical SCCmec element to that present in the Irish CC130 human MRSA. These researchers also identified other MRSA harbouring the novel SCCmec element emerging in bovine and human populations in the United Kingdom and Denmark. This study will be published simultaneously in Lancet Infectious Diseases.
Commenting on the significance of the findings, Professor Coleman said: “The results of our study and the independent United Kingdom study indicate that new types of MRSA that can colonise and infect humans are currently emerging from animal reservoirs in Ireland and elsewhere in Europe and that it is difficult to correctly identify them as MRSA. This knowledge will enable us to rapidly adapt existing genetic MRSA detection tests, but has also provided invaluable insights into the evolution and origins of MRSA.”
Co-authors on the Trinity study are Anna Shore, PhD; Emily Deasy, BA (Mod.); Peter Slickers, PhD, Grainne Brennan, MSc; Brian O’Connell, MD; Stefan Monecke, PhD; Ralf Ehricht, PhD; and David Coleman, PhD.
The manuscript for the study is being published ahead of print. The final, edited article is scheduled to be published in the August 2011 issue of the journal.
A.C. Shore, E.C. Deasy, P. Slickers, G. Brennan, B. O’Connell, S. Monecke, R. Ehricht, and D.C. Coleman. 2011. Detection of Staphylococcal Cassette Chromosome Mec Type XI encoding highly divergent mecA, mecI, mecR1, blaZ and ccr genes in human clinical clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 55:
