TCD Microbiologists Publish Genome Sequence of Infection Causing Staphylococcus Lugdunensis
Posted on: 11 July 2011
The genome sequence of Staphylococcus lugdunensis, an organism that can cause serious human infections in skin and soft tissue including heart valves, has recently been published by Trinity College scientists based in the School of Genetics and Microbiology in collaboration with researchers with the Sanger Institute at Hinxton, Cambridge. Their investigation into the precise role of the genes of S. lugdunensis could lead to better diagnostic and treatment strategies for infections associated with the bacteria.
Staphylococcus lugdunensis is a member of the genus Staphylococcus and is closely related to S. aureus, a leading cause of hospital-acquired infections. The Staphylococcus genus includes numerous species, most of which are harmless. However some can cause a wide variety of diseases and infections in humans. Infections caused by the S. lugdunensis organism, which resides on the surface of the skin, are currently under-reported because of its close similarity to S.aureus in diagnostic tests. By comparing their genomes and identifying the genetic differences specific to each organism, researchers were able to identify factors that are likely to be involved in bacteria’s colonisation of human skin and its ability to cause invasive infection.
One area of particular interest to the Trinity College scientists focuses on a set of genes, or locus, that encode the organism’s ability to extract much needed iron from haemoglobin in human blood, a crucial element in the bacteria’s ability to cause infection. The locus of S. lugdunensis is unusual in that it is capable of being amplified and experiments are currently underway to determine if the strains with extra copies of the locus have a growth advantage in the infected host compared to those with a single copy. This project underlines the diversity of Staphylococci that interact with humans and will inform studies to identify and characterise virulence and colonisation factors. By identifying specific gene locations, researchers can begin to develop strategies to prevent or cure infections associated with these pathogenic bacteria.
The Trinity team working on the project include Simon Heilbronner, a PhD student who is funded by an IRCSET scholarship, with assistance from Dr Joan Geoghegan and Dr Ian Monk, both postdoctoral researchers funded by Science Foundation Ireland. They work under the guidance of Professor of Molecular Microbiology at the School of Genetics and Microbiology, Tim Foster at the College’s Moyne Institute of Preventive Medicine. Genomic sequence analysis in the Sanger Institute was conducted by Dr Stephen Bentley, Dr Matthew Holden and Dr Julian Parkhill.
Professor Tim Foster is the lead inventor on several intellectual property patents that claim the use of surface proteins of S. aureus for therapeutic purposes. This intellectual property, which stemmed from research discoveries in Professor Fosters laboratory, was exclusively licensed by TCD to Inhibitex, a clinical stage biopharmaceutical company, in 1999 and further sub-licensed to Pfizer Vaccines in 2001. The latter are developing a vaccine for S. aureus infections which include Clumping Factor, a patent-protected protein. This vaccine has undergone a Phase I clinical trial for dose range and safety. A large scale Phase III trial to text efficacy is planned. More information regarding the product is available online.
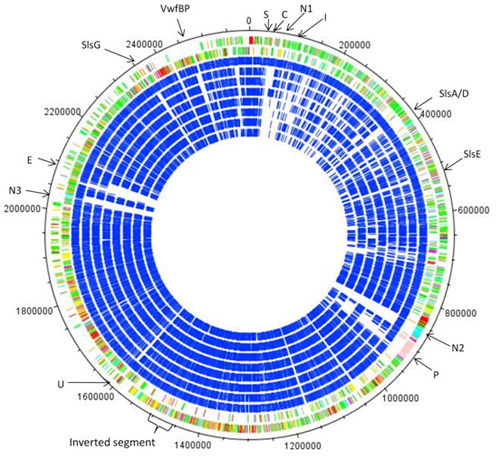
Diagram showing the genes encoded by the two DNA strands of the ~2,600,000 bp circular chromosome of S.lugdunensis N920143 (two outermost circles) and an overall comparison with S.lugdunesis HKU09-01 (outermost blue circle) followed by S.aureus and then other species of staphylococci. The arrows show the locations of putative virulence and colonisation factors. The gaps in the blue circles are S. lugdunensis sequences that are absent in other species.
